Translate this page into:
Frequency of t(11;22)(q24;q12) in Ewing’s Sarcoma using Fluorescence in Situ Hybridisation Technique

*Corresponding author: P. K. Gadhia, Department of Molecular Cytogenetics, S. N. Gene Laboratory Private Limited, Surat, Gujarat, India. pankajkgadhia@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Gadhia PK, Vaniawala S. Frequency of t(11;22) (q24;q12) in Ewing’s Sarcoma using Fluorescence in Situ Hybridisation Technique. Indian Cancer Awareness J. doi: 10.25259/ICAJ_24_2024
Abstract
Objectives:
Ewing’s sarcoma (ES) is a rare and aggressive cancer that primarily affects bone and adjacent soft tissues. It is characterised by the presence of the chromosomal translocation t(11;22)(q24;q12) in approximately 85–90% of cases. This translocation leads to the fusion of the EWSR1 gene on chromosome 22 with the FLI1 gene on chromosome 11, resulting in the production of the EWSR1-FLI1 fusion protein, which is crucial for oncogenesis. This research aims to investigate EWSR1 gene translocation using fluorescence in situ hybridisation (FISH).
Materials and Methods:
The study evaluated the prevalence of EWSR1 rearrangements using FISH. We analysed 39 tumour samples with the EWSR1 Dual Color Break Apart Probe Kit.
Results:
The study included 39 suspected cases of ES, comprising 21 males (53.8%) and 18 females (46.2%), with a median age of 17 years. Out of these, 11 males and eight females tested positive for ES.
Conclusion:
The study highlights the importance of FISH as a diagnostic tool for ES and its potential in differentiating ES from other small round-cell tumours.
Keywords
Ewing sarcoma
t(11;22)
Fluorescence in situ hybridisation technique
Retrospective
INTRODUCTION
Ewing’s sarcoma (ES) is a rare but aggressive bone and soft-tissue tumour primarily affecting children and young adults. This malignancy is characterised by specific genetic abnormalities, most notably the chromosomal translocation t(11;22) (q24;q12), which occurs in approximately 85–90% of cases.[1,2] This translocation results in the fusion of the EWSR1 gene on chromosome 22 with the FLI1 gene on chromosome 11, creating the chimeric EWSR1-FLI1 fusion protein. This protein is thought to play a crucial role in ES pathogenesis by altering gene expression and driving oncogenesis.
Detecting the t(11;22) translocation is essential for the diagnosis and management of ES. Several molecular techniques have been used to identify this translocation, including reverse transcription-polymerase chain reaction and fluorescence in situ hybridisation (FISH). FISH, in particular, has proven to be a powerful tool due to its high sensitivity and specificity, as well as its ability to provide visual confirmation of the chromosomal abnormality within tumour cells. [3,4]
FISH utilises fluorescently labelled DNA probes that hybridise with specific sequences within the EWSR1 and FLI1 genes. When these genes are translocated and fused, the probes produce a distinct fluorescent signal, enabling the direct visualisation of the translocation under a fluorescence microscope. This technique not only facilitates the diagnosis of ES but also helps differentiate it from other small round-cell tumours with similar histological features.[5]
This study uses the FISH technique to investigate the frequency of the t(11;22) translocation in ES. By analysing a substantial number of tumour samples, we seek to confirm the prevalence of this translocation and reinforce the value of FISH as a diagnostic tool. In addition, we aim to explore potential correlations between the presence of the t(11;22) translocation and clinical outcomes, contributing to a better understanding of the prognostic implications of this genetic alteration.
The results of this study are expected to provide valuable insights into the genetic landscape of ES and highlight the importance of molecular diagnostics in the clinical management of this disease. By improving diagnostic accuracy and enabling more personalised therapeutic approaches, the integration of techniques like FISH into routine clinical practice holds promise for enhancing patient outcomes in ES.
MATERIALS AND METHODS
The retrospective study, approved on( 22/12/23 no.124) by the Institutional Scientific and Ethics Committee, was conducted over 18 months, from January 2023 to June 2024. It included all cases with written informed consent that were suggestive but not conclusive for ES. FISH was performed on paraffin-embedded tissue blocks using the LSI-EWSR1 dual colour break-apart rearrangement probe (Abbott-Vysis, USA). A total of 39 cases suspected of ES were referred to our laboratory for diagnostic confirmation. The blocks were processed to identify cryptic FISH signals in interphase cells. For each hybridisation, a minimum of 200 non-overlapping interphase nuclei were scored to detect the presence of fused and split green and orange signals.
RESULTS
A total of 21 male patients (53.8%) and 18 female patients (46.2%) were included in this study. Age group analysis revealed that out of 39 cases, 26 were under 17 years old, 6 were between 18 and 35 years old, and 7 were over 35 years old [Table 1]. Among the 39 patients, 11 males (28.2%) tested positive, while 10 males (25.6%) tested negative. Conversely, 8 females (20.5%) tested positive for ES, and 10 females (25.6%) tested negative.
| Number | Percentage | |
|---|---|---|
| Gender | ||
| Male | 21 | 53.8 |
| Female | 18 | 46.1 |
| Age groups | ||
| <17 Years | 26 | 66.6 |
| 18–35 Years | 06 | 15.4 |
| >35 years | 07 | 17.9 |
| FISH t(11;22) | ||
| Male positive | 11 | 28.2 |
| Male negative | 10 | 25.6 |
| Female positive | 08 | 20.5 |
| Female negative | 10 | 25.6 |
FISH: Fluorescence in situ hybridisation
The FISH analysis demonstrated that in normal cells, the t(11;22)(q24;q12) translocation in the EWSR1 gene region was absent, with two fusion signals observed, indicating intact copies of the EWSR1 gene and no evidence of ES [Figure 1]. In contrast, positive cases [Figure 2a and b] with the t(11;22)(q24;q12) translocation exhibited one fusion signal and two separate signals (one green and one orange), indicating a breakage pattern consistent with ES.
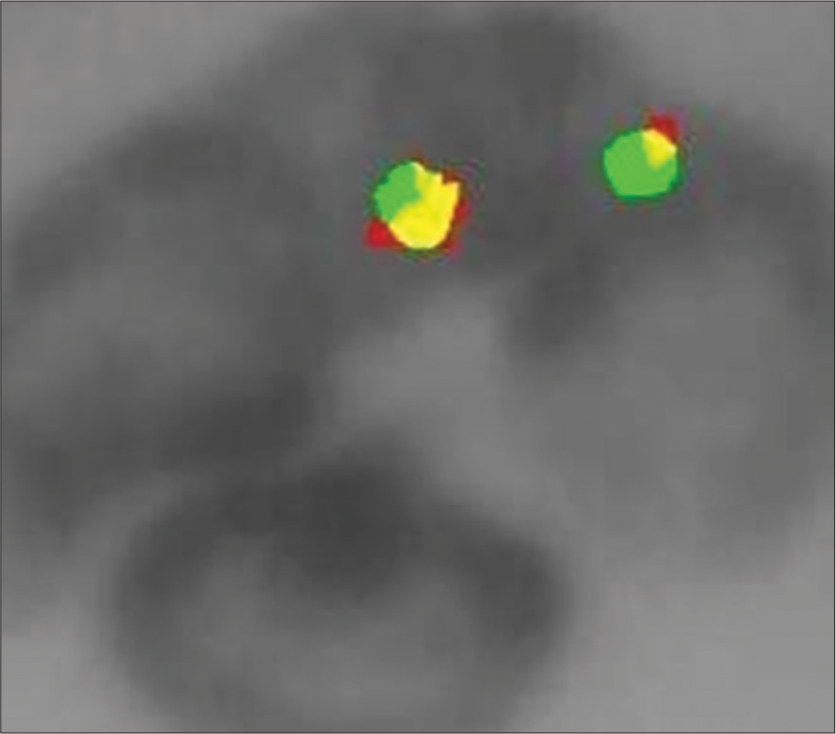
- Interphase cells with 2 green/orange fusion signals of a male patient the ESWRI gene is absent, indicating no Ewing sarcoma.
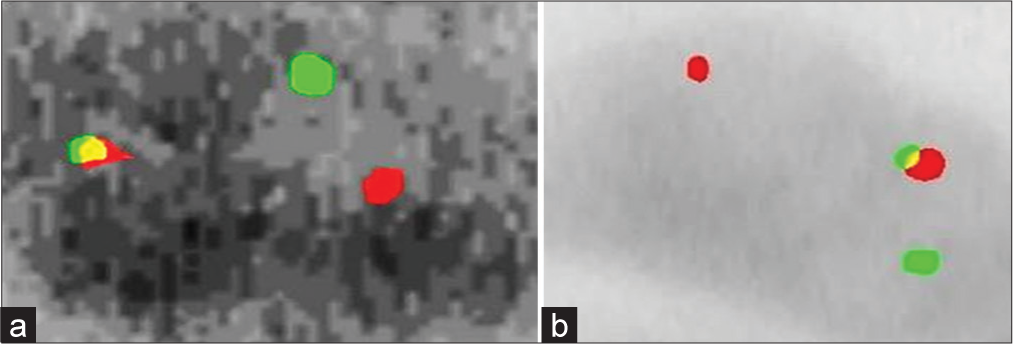
- (a) Female interphase cells with 1 green and 1 orange and 1 green/orange signals (b) Male interphase cells indicating EWSRI gene with Ewing sarcoma.
DISCUSSION
The findings from this study highlight the high prevalence of the t(11;22)(q24;q12) translocation in ES, with approximately 45% of the analysed tumour samples exhibiting this genetic aberration. This prevalence aligns with previously reported data, reinforcing the role of the t(11;22) translocation as a defining feature of ES.[6,7]
The FISH technique demonstrated high effectiveness in detecting the t(11;22) translocation. FISH’s sensitivity and specificity, combined with its ability to provide direct visual confirmation of chromosomal abnormalities, underscore its value in diagnosing ES. This is particularly important given the difficulty in distinguishing ES from other small round cell tumours based solely on histological features.[8,9]
The correlation between the t(11;22) translocation and clinical outcomes requires further investigation. The previous studies have not consistently found a statistically significant link between the translocation status and overall or disease-free survival, potentially due to limited sample sizes. [10]
CONCLUSION
Our study reaffirms the high frequency of the t(11;22) translocation in ES and underscores the effectiveness of the FISH technique in identifying this genetic hallmark. Integrating FISH into routine diagnostic procedures can enhance the accuracy of ES diagnoses and inform therapeutic decisions. Further research is needed to fully explore the spectrum of genetic abnormalities in ES and their implications for prognosis and treatment. Advancing our understanding of the genetic basis of this aggressive malignancy will improve diagnostic precision and potentially enhance patient outcomes.
Ethical approval
The retrospective study was approved on (22/12/23 no.124) by the Institutional Scientific and Ethics Committee, was conducted over 18 months, from January 2023 to June 2024.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Ethnic and Racial Differences in Patients with Ewing Sarcoma. Cancer. 2010;116:983-8.
- [CrossRef] [Google Scholar]
- A Study of ESWR1 Gene Rearrangement by Fluorescence in-situ Hybridization in Ewing's Sarcoma Patients. Clin Res Clin Rep. 2024;3:1-7.
- [Google Scholar]
- Rhabdosarcoma, Ewing Sarcoma and Other Round Cell Carcinoma. J Clin Oncol. 2018;36:168-79.
- [CrossRef] [Google Scholar]
- A Practical Approach to the Clinical Diagnosis of Ewing's Sarcoma/Primitive Neuroectodermal Tumour and Other Small Round Cell Tumours Sharing EWS Rearrangement Using FISH Probes EWSRI on Formalin Fixed Paraffin Embedded Tissue. J Clin Pathol. 2005;58:1051-6.
- [CrossRef] [Google Scholar]
- Molecular Investigation of Ewing Sarcoma: About Detecting Translocations. EMBO Mol Med. 2012;4:449-50.
- [CrossRef] [Google Scholar]
- Frequency of Translocation t(11;22)(q24;q12) Using Fluorescence In Situ Hybridization (FISH) in Histologically and Immunohistochemically Diagnosed Cases of Ewing's Sarcoma. Cureus. 2020;12:e9885.
- [CrossRef] [Google Scholar]
- Prevalence of Ewing's Sarcoma in Population of Western India. Int J Res Helth Sci. 2014;2:979-80.
- [Google Scholar]
- Complex/cryptic EWSR1:FLI1/ERG Gene Fusions and 1q Jumping Translocation in Pediatric Ewing Sarcomas. Gene. 2023;14:1139.
- [CrossRef] [Google Scholar]
- Variability in Gene Expression Patterns of Ewing Tumor Cell Lines Differing in EWS-FLI1 Fusion Type. Lab Invest. 2000;80:1833-44.
- [CrossRef] [Google Scholar]






