Translate this page into:
Next-generation Sequencing: For the Present Generation Oncologist

*Corresponding author: Abhishek Pathak, Command hospital, Kolkata, India. drabhipat@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Gupta A, Pathak A, Ranjan R, Bandyopadhyay A, Rathore A. Next-generation sequencing: For the present generation oncologist. Indian Cancer Awareness J 2022;1:2-7.
Abstract
Previously, doctors had to treat this deadly disease based on immunohistochemistry, polymerase chain reaction and fluorescence in situ hybridisation-based diagnostics tests information which used to provide limited information. There was always a requirement of a single test/assay which could give all these information in a single assay and without much loss of precious sample. Comprehensive next-generation sequencing (NGS)-based assays are gaining much importance in treatment decision among clinicians. These present technologies using DNA, RNA, and methylation sequencing have brought a lot of changes in cancer therapeutics. Oncogenic drivers are genomic alteration lead to malignant transformation and progression of cancer. The interpretation of the results of NGS is also very challenging as the results obtained are ‘variants’ are of unknown prognostic significance. Apart from these, NGS helps us in documenting various other genomic signatures such as tumour mutation burden and microsatellite instability. There are now multiple gene panels that are recommended by major international societies such as NCCN, for the treatment of various malignancies. In fact, NGS forms the most important pillar of precision oncology and has brought a paradigm shift in the way major cancers are treated. It is still an evolving field and many times the interpretation of the results of NGS reports is very difficult. NGS-based precision medicine treatment offers true value addition in clinical practice has a positive impact on patient lives in cases of refractory cancer, the role of NGS is the only source of light that helps us in navigating amidst the darkness and hopelessness of a refractory malignancy. In fact, presently not only in refractory malignancies but the role of NGS has come in the management of cancer patients in the first line as it gives us a more comprehensive understanding of the disease. This review article has been written with an idea to make a general practitioner aware of this novel technique, the advantages as well as the pitfalls.
Keywords
NGS
PCR
FISH
INTRODUCTION
Cancer is a deadly disease and a nightmare for every patient and their family. Over time, the diagnosis of cancer has seen a paradigm shift with newer diagnostic modalities emerging both in laboratory sciences and radiological sciences. Previously, doctors had to treat this deadly disease based on immunohistochemistry (IHC), polymerase chain reaction (PCR) and fluorescence in situ hybridisation (FISH)-based diagnostics tests information which used to provide limited information. However, now, the outlook is getting changed as clinicians have an unmet need to get all the biomarker-based information more comprehensively in a single test or preferably in one or two tests. This comprehensive test will help the clinician to get all the guidelines based on genetic alteration and genetic signatures information which eventually opens up the broad possibility to treat their patient with combination therapy. There was always a requirement of a single test/assay which could give all this information in a single assay and without much loss of precious sample.
Comprehensive next-generation sequencing (NGS)-based assays are gaining much importance in treatment decisions among clinicians. These present technologies using DNA, RNA, and methylation sequencing have brought a lot of changes in cancer therapeutics. Oncogenic drivers are genomic alteration lead to malignant transformation and progression of cancer.[1] NGS is a type of DNA sequencing technology that uses parallel sequencing of multiple small fragments of DNA to determine the sequence. This NGS uses a ‘high-throughput’ technology which increases the speed at which an individual’s genome can be sequenced. NGS interrogates all the known and unknown genetic alterations and genomic signature in one test with less time keeping the clinicians’ headache at bay for opting for different tests one after another from different diagnostics providers.
FIRST-GENERATION SEQUENCING – THE HISTORY
The first-generation sequencing was developed by 2 times Nobel laureate Frederick Sanger and his colleagues Maxam and Gilbert in 1977.[2] Hence, the name Sanger sequencing. It is also commonly referred to as ‘conventional’ or ‘traditional’ sequencing.
It determines the sequence of large DNA fragments (up to approximately 500–900 bases), using radioactive markers for each nucleotide, and later adaptations have used fluorescently tagged versions. It is a costly and time taking procedure. There has been some prediction that sequencing the entire human genome will take around 60 years.[3]
WHAT IS NGS?
NGS is the technology used for determining the order of nucleotide sequence in DNA and RNA and to study genetic variation associated with diseases or other biological phenomena. Previously, the clinician had to rely on single gene-based tests, then came hotspots based sequencing tests. Now, as the demand for genomic information is increasing and sequencing, the cost is reducing, clinicians are getting more inclined toward adopting multigene assay and more precise comprehensive genomic tests for target-based therapies.
For any sequencing to be done, the first step is in recovering nucleic acid. The most common source for which is the formalin-fixed paraffin-embedded (FFPE) tumour tissue. The basic next-generation sequencing process involves fragmenting DNA/RNA into multiple pieces and then adding adapters subsequently sequencing the libraries and reassembling them to form a genomic sequence. NGS sequences millions of fragments in a parallel fashion, improving speed and accuracy while reducing the cost of sequencing. Next-generation sequencing involves three basic steps: Library preparation, sequencing, and data analysis [Graph 1]. A fresh sample is always preferred as the nucleic acid in archival samples is degraded. Other sources of nucleic acids are cfDNA/ctDNA, the so-called liquid biopsies. These allow an indirect genomic analysis of tumour by targeted amplicon deep sequencing or by digital PCR.[4]
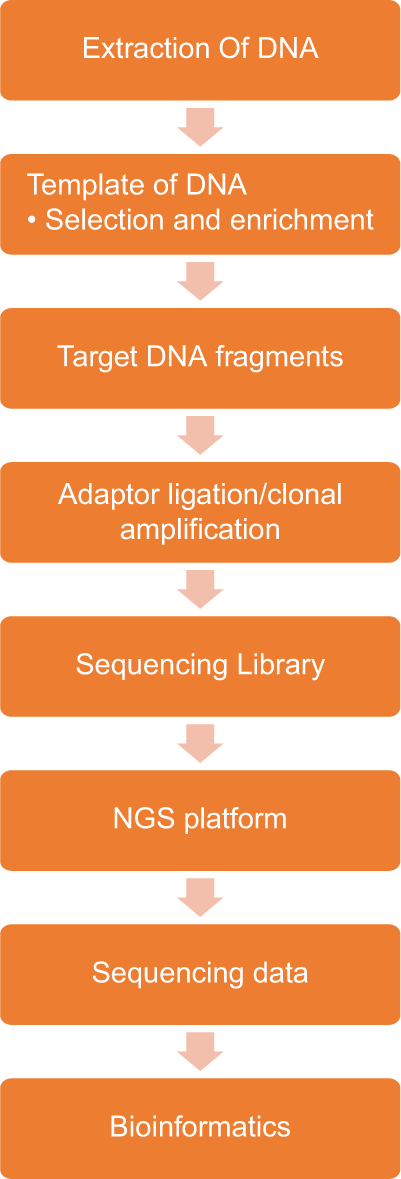
- Next-generation sequencing working chart.
WHAT ARE PLATFORMS FOR NGS?
‘Platforms’ are sequencing instruments along with their reagents. These platforms prepare ‘Library’ by purifying patients’ DNA, amplifying it, and then physical isolation of DNA fragments by attaching them to solid surfaces or beads. These data are electronically generated and computationally aligned against ‘reference genome’ or sequence. The reference genome GRCh 38 was released by the National Centre of Biotechnology Innovation on December 2013.
The third-generation sequencing is a further refined technique of sequencing where single DNA fragments are used without amplifying, thereby reducing the chances of any error as they do not use any amplification process. A few of the commonly used platforms are given below.
| NGS platforms/company/max output per run | Read length per run (bp) | Platform cost (USD approx.) |
|---|---|---|
| GS Junior/Roche/70 Mb | 500 | 100,000 |
| HiSeq/Illumina/1500 Gb | 2×150 | 750,000 |
| MiSeq/Illumina/15 Gb | 2×300 | 125,000 |
| SOLiD/Life Technologies/120 Gb | 50 | 350,000 |
GENOMIC ALTERATION
The various genomic alterations which are detected by NGS include
Chromosomal structural variants such as translocation/ insertion/inversions (detected by WGS)
Gene fusions – Gain of functions
Gene fusion – loss of functions
Zygosity and copy number alteration
Chromosomal instability
Tumour mutational burden.
WHAT ARE DIFFERENT TYPES OF SEQUENCING?
Depending on the extent of DNA sequencing, there are whole-genome sequencing or limited portions of DNA sequencing such as whole-exome sequencing or targeted panel sequencing. Whole-genome sequencing involves 3.3 × 109 bases sequencing. It is a costly affair compared to exome sequencing or targeted panel sequencing. It sequences both the coding and non-coding DNA, the role of which presently is not understood. Exome sequencing is done for the segment of DNA which is known to translate into proteins. This accounts for only 2% of the entire genome. This is a cheaper alternative and almost 85% of the diseases have known mutations in the exome. The ideal way would be to go for exome sequencing initially, and in case, it is non-diagnostic then whole-genome sequencing can be tried. Targeted gene panels are used to evaluate a set of genes known to be involved in a particular disease. These are less costly than whole-exome sequencing and have a greater sensitivity and depth.
HOW TO INTERPRET THE RESULTS?
The interpretation of results of NGS is also very challenging as the results obtained are ‘variants’ are of unknown prognostic significance.[5] Results obtained are mainly as one of the follow
‘Pathogenic’ – these are variants that have been previously reported in patients with the disease or are strongly suspected of being pathogenic based on preclinical studies
‘Likely pathogenic’ – these are sequences that are likely to be implicated in disease pathology
‘Likely benign’ – sequences wherein the majority of evidence suggests the effect of the variant is benign
‘Benign’ – sequences which do not alter gene expression or function
‘Unknown clinical significance’ also called ‘variant of uncertain significance’ or ‘finding of unknown clinical significance’ is sequences for which neither their benign nor malignant potential is clear.
Clinical NGS laboratories generally will be in agreement on their interpretation of a variant as pathogenic or benign.
WHAT IS SOMATIC AND GERMLINE MUTATION?
In respect to the cell of origin, cancer can be termed as germline and somatic. Germline as the name suggests occurs in our germ cells and is passed on from generation to generation and is hereditary. Whereas, somatic/acquired mutations are the effect of environmental factors, lifestyle, etc. Somatic mutations are not hereditary, and one individual acquires these mutations in their lifetime. Germline mutation-causing cancers are quite rare in the population like 5–10% in the case of breast cancer and somatic cancers are 95–90%.[6] Somatic mutations are present only in tumour cells whereas, in Germline, its presence is in all the cells of our body.
OTHER GENETIC SIGNATURES DETECTED ON NGS
Nowadays, different drugs have been approved to target genetic variation causing cancer, but those drugs are not approved for all types of cancer. With the help of NGS data, it is documented that different genomic signatures have promising therapeutic significance in cancer management and they are tissue agnostic.
Tumour mutation burden (TMB) and microsatellite instability (MSI) are two genomic signatures that different guidelines have recommended for the treatment of tissue agnostic solid tumours.
TMB is defined as the total number of somatic mutations per coding area of a tumour genome is an emerging clinical biomarker associated with response to immune checkpoint inhibitor (ICI) therapy in lung cancer, melanoma, gastric cancer, and urothelial cancer.[7,8] Whole-exome sequencing TMB is the gold standard. However, due to its high cost targeted and long turnaround time targeted, sequencing based NGS has gained more popularity.[9] Food and Drug Administration (FDA) approved FoundationOne CDx and MSK-IMPACT panel for assessing TMB.
Whereas, MSI is checked to know whether DNA mismatch repair genes are functioning properly or not. DNA mismatch repair genes play a critical role in maintaining genomic stability by repairing genetic mutation occurring more frequently in the microsatellite region of our genome during the replication process. Hence, it is evidenced that with the help of NGS, TMB- and MSI-based treatment planning can be administered, along with targetable mutated genetic information in the clinical setting. To provide a complete outlook, MSI and TMB should be done on whole-exome based sequencing as it is proved to be the gold standard.
It is clear now that clinicians can get all the relevant information not only on genetic variation but also on genomic signatures as per guidelines in a single assay, thus saving time, cost, and loss of precious sample loss.
In one retrospective study, it has been found that patient treated with NGS-based precision medicine treatment has shown an increase in progression-free survival (PFS) than those who are treated with non-NGS-based precision medicine-based therapy.
CLINICAL USE OF NGS
Hence, when should a clinician ask for NGS? It is not one of the routine investigations wherein every patient who visits an oncologist is bound to get it done. Besides, it is an expensive and time-consuming procedure. Hence, the main role of NGS comes when we are considering a large number of pathogenic genes and there is a high suspicion of family/genetic basis.[10]
In oncological practice, NGS has been used in screening, diagnosing, and also in planning the treatment. There are now multiple gene panels that are recommended by major international societies such as NCCN, for the treatment of various malignancies.[11] Few societies such as the American Society of Breast Surgeons have recommended all patients of breast cancer to undergo testing for breast cancer susceptibility genes 1 (BRCA1), BRCA2, and partner and localiser of BRCA2 (PALB2), these recommendations were based on an article from Beitsch et al.[12]
Multigene panels including BRCA 1 and 2, ATM, CDH1, NF1, PALB2, and TP53 are used in patients with a history of breast, ovarian, prostatic, and even pancreatic cancers. Various genes which are studied in gastrointestinal malignancies are APC, BMPR1A, EPCAM, MLH1, MSH2, MSH6, MYUTH, PMS2, PTEN, SMAD4, CHEK2, POLD1, TP53, etc. Similarly, we have genetic abnormalities in haematological malignancies AML and ALL. The US FDA had approved two tests by name MSK-IMPACT and F1CDx for detecting pathogenic mutations in solid tumours using FFPE tissues.[13] These tests detect variations in the coding region of over 400 genes and other genomic signatures such as MSI and tumour mutational burden.
The mutations can be detected in the germline as well as somatic mutations which are present in tumour cells/circulating DNA. There have been studies such as NCI-MATCH wherein the use of pembrolizumab was based on MSI/dMMR report leading to a paradigm shift into the treatment of cancer based on genetic abnormality rather than based on tissue of origin.[14]
CLINICAL TRIALS ASSOCIATED WITH NGS
Various clinical trials have been conducted to prove the efficacy of NGS-based treatment. In earlier trials such as SAFIR01 trial, almost 30% of patients had shown objective response or stable disease in ‘hard to treat cancers.’[15]
In SHIVA randomised trials, there was no improvement in PFS in patients treated on the basis of NGS versus standard of care.[16] However, in MOSCATO 01 trial, we have shown that molecular screening strategies led to a matched targeted treatment in 19% of successfully screened patients, and 7% of the successfully screened obtained a PFS ratio above the predefined threshold.[17]
PRECISION ONCOLOGY
As cancer treatment is evolving on daily basis, treatment is becoming more personalised. As it has been documented that two individuals with the same tumour cannot avail same treatment regime as their tumour biology differs to great extent. Based on this, clinician prefers to treat their patient in a more precise way rather than previously practiced conventional therapeutic approach. This is also termed as ‘Precision Medicine.’
Precision oncology is a field of medicine wherein the genomic and protein expression signature of a patients’ tumour tissue along with the clinical and pathological presentation is used for accurate diagnosis and plan treatment with maximum benefits and with least side effects. Precision medicine is one of the approaches in clinical practice, where the information about an individual’s genetic information ‘blueprint’ or protein signature in combination with the clinical signs and symptoms of presentation is used to diagnose, prevent or treat a disease condition.
HOW HAS THE TREATMENT LANDSCAPE CHANGED DUE TO NGS?
Pembrolizumab leading to significant longer PFS in the first line than chemotherapy as first-line treatment in metastatic colorectal cancer with MSI-H/dMMR tumours (KEYNOTE -177)[18]
Larotrectinib for solid tumours with NTRK gene fusions (LOXO-TRK-14001 (NCT02122913), SCOUT (NCT02637687), and NAVIGATE (NCT02576431)
NRG 1 fusion in lung cancer being treated with afatinib[19,20] NRG1 fusions may be oncogenic, and oncogenic fusions of NRG1 may cause persistent activation of ErbB receptor tyrosine kinases that drive excess activity in the mTOR and MAPK pathways and promote tumourigenesis, the investigators noted
FDA approves pralsetinib for RET mutant thyroid cancer.
HOW DOES NGS PROVE A BETTER DIAGNOSTIC TOOL FOR BETTER CANCER MANAGEMENT?
Saves tissue and time compared to sequential testing
Helps to make clinical decisions based on the molecular spectrum as it covers all the biomarkers given in the guidelines, as well as other novel biomarkers information which is currently in clinical trials
It proves a potential tool to provide clinical trial alternatives to patients when FDA approved drugs are not an option
Helps oncologists to eliminate futile or potentially harmful treatment based on the patients’ germline and tumour genetic signature
It has pan-cancer coverage
ADVANTAGES AND DISADVANTAGES OF NGS
The advantages of this technique are many such as
It is high throughput can sequence multiple samples at a given time
Highly sensitive and specific and offers a single-nucleotide resolution
Can screen multiple genes in multiple samples simultaneously
Gives a better picture of tumour heterogeneity
Constantly improving technology
Confers decreased sequencing costs per gene.
However, there are a few disadvantages of the same
Strong analytical and bioinformatics teams are required
The identified alteration may not have clinical significance as of now
Requires reflex testing of some of the larger variations to confirm
Comparatively costly and time taking than IHC, PCR, and FISH for known variant detection.
CONCLUSION
NGS-based precision medicine treatment offers true value addition in clinical practice has a positive impact on patient lives. In cases of refractory cancer, the role of NGS is the only source of light that helps us in navigating amidst the darkness and hopelessness of a refractory malignancy.
The most common source of extaracting nucleic acid is formalin fixed paraffin embedded (FFPE) blocks.
NGS sequences millions of fragments together reducing cost and time of sequencing.
Sanger sequencing was named after Frederick Sanger.
NGS helps us in deeper understanding of the disease and helps us in deciding the first line and subsequent therapy.
NGS provides a paradigm shift in the treatment decision and better management of cancer patients.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- The genetic basis for cancer treatment decisions. Cell. 2012;148:409-20.
- [CrossRef] [PubMed] [Google Scholar]
- Key principles and clinical applications of “next-generation” DNA sequencing. Cancer Prev Res (Phila). 2012;5:887-900.
- [CrossRef] [PubMed] [Google Scholar]
- Toward the 1, 000 dollars human genome. Pharmacogenomics. 2005;6:373-82.
- [CrossRef] [PubMed] [Google Scholar]
- Next-Generation sequencing to diagnose suspected genetic disorders. N Engl J Med. 2018;379:1353-62.
- [CrossRef] [PubMed] [Google Scholar]
- Available from: https://www.nccn.org/professionals/physician_gls/default.aspx [Last assessed on 2021 May 20]
- Underdiagnosis of hereditary breast cancer: Are genetic testing guidelines a tool or an obstacle? J Clin Oncol. 2019;37:453.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of Automatic Class III Designation for MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) 2021. Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/den170058.pdf. [Last accessed on 2020 May 22]
- [Google Scholar]
- Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/125514s024lbl.pdf [Last assessed on 2021 May 20]
- Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet. 2015;47:717-26.
- [CrossRef] [PubMed] [Google Scholar]
- Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124-8.
- [CrossRef] [PubMed] [Google Scholar]
- Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207-11.
- [CrossRef] [PubMed] [Google Scholar]
- Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909-20.
- [CrossRef] [Google Scholar]
- Implementing tumor mutational burden (TMB) analysis in routine diagnostics-a primer for molecular pathologists and clinicians. Transl Lung Cancer Res. 2018;7:703-15.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: A multicentre, prospective trial (SAFIR01/ UNICANCER) Lancet Oncol. 2014;15:267-74.
- [CrossRef] [Google Scholar]
- Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324-34.
- [CrossRef] [Google Scholar]
- High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: Results of the MOSCATO 01 trial. Cancer Discov. 2017;7:586-95.
- [CrossRef] [PubMed] [Google Scholar]
- Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383:2207-18.
- [CrossRef] [PubMed] [Google Scholar]
- Pembrolizumab in MSI-H-dMMR advanced colorectal cancer-a new standard of care. N Engl J Med. 2020;383:2283-5.
- [CrossRef] [PubMed] [Google Scholar]
- A Real-World Feasibility Study of Patients with Solid Tumors Harboring NRG1 Gene Fusions: NSCLC Subset Analysis. Presented At: 2020 International Association for the Study of Lung Cancer. 2021
- [CrossRef] [Google Scholar]






